Here's my poster for #ABACBS2023!
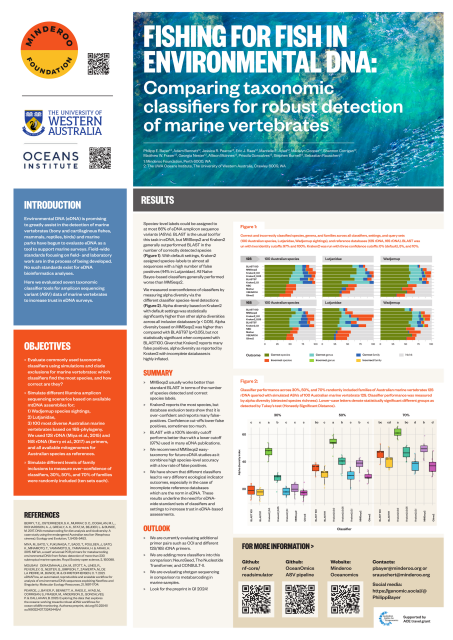
We compared different #eDNA taxonomic classifiers across different simulated datasets of 12S and 16S metabarcoding data.
MMSeqs2 generally 'wins' closely followed by BLAST.
With exclusion databases it becomes apparent that standard Kraken2 produces many, many false positives. With some families like snappers, all classifiers can't assign more than ~30% of sequences.
(not shown is COI, where MMSeqs2 and Mothur are working best)
Frank Aylward
in reply to Philipp Bayer • • •Philipp Bayer
in reply to Frank Aylward • • •I sent them an initial outline in powerpoint and my ggplot figures, we had a few calls and sits together. they redrew the figures in illustrator to make them nice! the text is relatively unchanged, which is why there's so much of it 😀
Frank Aylward
in reply to Philipp Bayer • • •