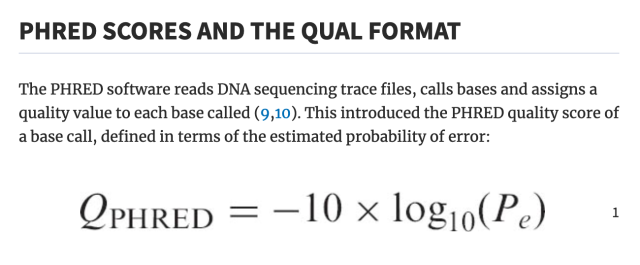
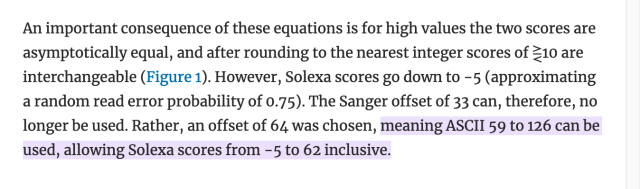
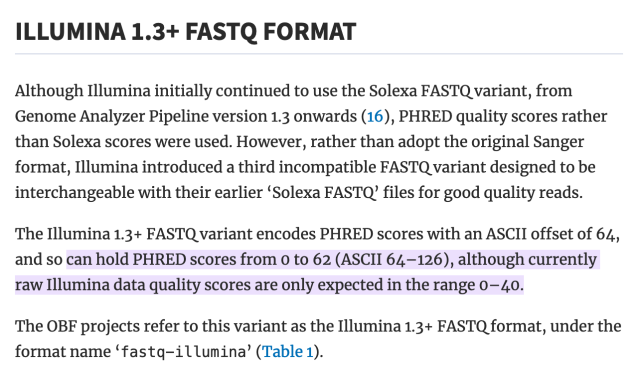
literally who hurt genomics to make you all encode one specific kind of number as the ASCII characters from ! to ~ as an integer input to some logarithm function, but then others of you changed the function but kept encoding it as a single ASCII character ranging from -5 to 62 (???), and then later they decided that -5 to 62 was silly and so they changed that to 0-62 and throwing away half the original range for no reason, except actually it's 0-40 by convention.
did anyone consider "encoding it as a number"

Dr. Angus Andrea Grieve-Smith
in reply to jonny (good kind) • • •Eli Roberson (he/him)
in reply to Dr. Angus Andrea Grieve-Smith • • •@grvsmth filesize advantage. Quality as a number would require minimum two characters (single digit plus delimiter) and maximum of 3 characters (two numbers plus delimiter). Fixed width 0 led would still require two characters. ASCII coding makes it one character per quality score. Minimizes cold file sizes.
Now the multiple FASTQ quality ASCII coding specs are an artifact of Illumina choices. Sanger phred scale should have been the standard all along.