Search
Items tagged with: Genomics
Hot off the press! Our latest work on the evolution of facultative symbiosis in stony corals, focusing on a remarkable Mediterranean species: Oculina patagonica.
🪸 🌊
#evobio #corals #coralbiology #genomics
nature.com/articles/s41586-025…
The evolution of facultative symbiosis in stony corals - Nature
Genomic sequencing of the thermotolerant coral species Oculina patagonica, single-cell transcriptomic analyses of symbiotic and non-symbiotic specimens and comparisons with obligate symbiotic coral species reveal adaptations that provide resilience t…Nature
Prevalence and Dynamics of Genome Rearrangements in Bacteria and Archaea
biorxiv.org/content/10.1101/20…
#genomics #bacteria #archaea #evolution

Prevalence and Dynamics of Genome Rearrangements in Bacteria and Archaea
The genetic material of bacteria and archaea is organized into various structures and set-ups, attesting that genome architecture is dynamic in these organisms.bioRxiv
Giant viruses integrate into the genomes of money protists. Our latest study adds the model protist Euglena to the list
"Giant endogenous viral elements in the genome of the model protist Euglena gracilis reveal past interactions with giant viruses"
biorxiv.org/content/10.1101/20…
#viruses #protists #microbiology #genomics

Giant endogenous viral elements in the genome of the model protist Euglena gracilis reveal past interactions with giant viruses
Giant viruses in the phylum Nucleocytoviricota have increasingly been found integrated into the genomes of diverse eukaryotes. Here we report 8 Giant Endogenous Viral Elements (GEVEs) in the genome of the microalgae Euglena gracilis.bioRxiv
Excited to announce our article, "EvoWeaver: large-scale prediction of gene functional associations from coevolutionary signals" was just published in Nature Communications!
We demonstrate that it's possible to infer how genes work together using only sequencing data by leveraging correlated signals of evolution. Check it out at the link below!
#genomics #bioinformatics #rstats #genetics #evolution #biology
nature.com/articles/s41467-025…
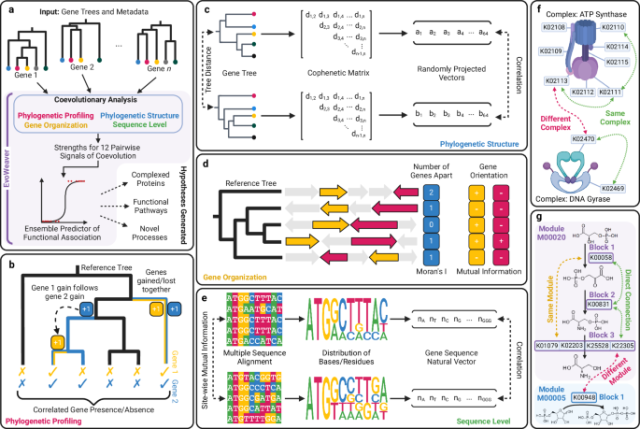
EvoWeaver: large-scale prediction of gene functional associations from coevolutionary signals - Nature Communications
Despite having structures for most proteins, we still do not know their function. Here, authors introduce EvoWeaver, a de novo method to identify genes working together using evolutionary information.Nature

Comparative genomics provides insights into chromosomal evolution and immunological adaptation in horseshoe bats - Nature Ecology & Evolution
A comparative analysis of bat genomes, including five newly assembled chromosome-scale genomes, provides insights into the diversification of horseshoe bats and reveals molecular adaptations with a potential role in antiviral immune response.Nature
Modeling the mosaic structure of bacterial genomes to infer their evolutionary history
#evolution #genomics #bacteria #bioinformatics #science
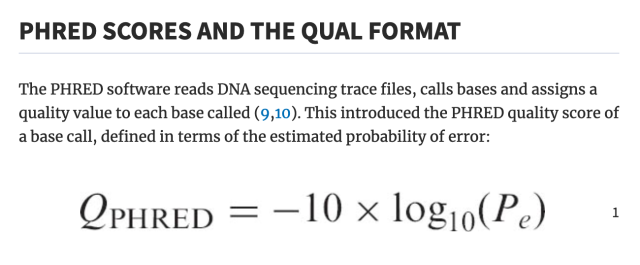
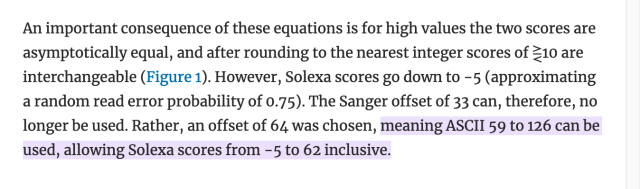
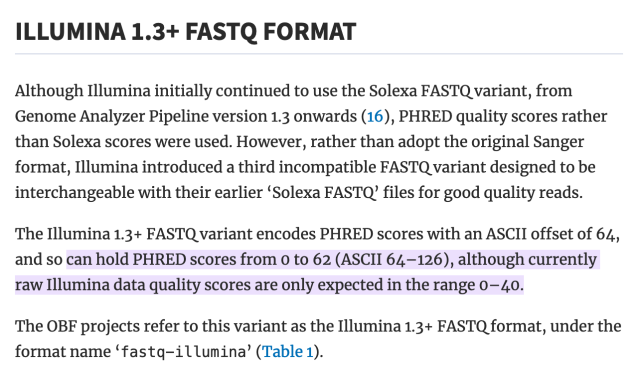
literally who hurt genomics to make you all encode one specific kind of number as the ASCII characters from ! to ~ as an integer input to some logarithm function, but then others of you changed the function but kept encoding it as a single ASCII character ranging from -5 to 62 (???), and then later they decided that -5 to 62 was silly and so they changed that to 0-62 and throwing away half the original range for no reason, except actually it's 0-40 by convention.
did anyone consider "encoding it as a number"
ORA is a type of enrichment analysis that analyses over-represented functional categories in gene lists. These tools have accumulated ~190k citations, but they have subtly different behaviours. Here we unpack the differences and investigate two subtle problems in some implementations, which may have negatively impacted those 190k research papers.
doi.org/10.1093/bioadv/vbae159
#genomics #bioinformatics
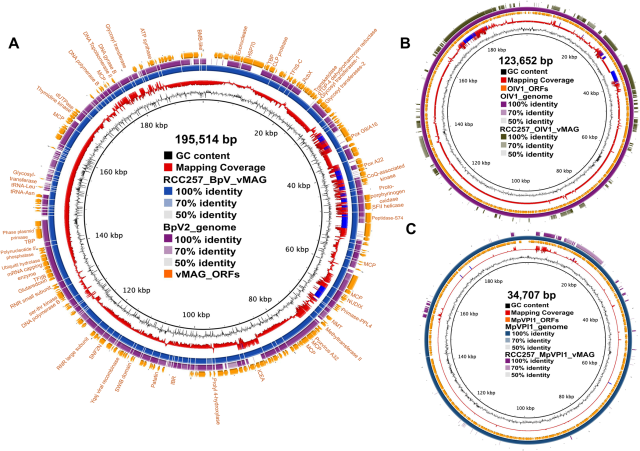
Genomic analyses of Symbiomonas scintillans show no evidence for endosymbiotic bacteria but does reveal the presence of giant viruses
Author summary Endosymbiotic bacteria are found in a wide variety of hosts across the tree of eukaryotes and have been proposed to be evolutionarily and ecologically significant, but in most cases, we know little to nothing about them.journals.plos.org
PhD studentship to characterise aphid immune evolution. Lots to uncover in an insect group with major ecological & economic impact. Aphid systemic immunity is a black box. The interested PhD candidate will both provide one of the first detailed descriptions of aphid immunity, but also uncover principles of immune evolution.
Deadline July 29th at #UoExeter
Email or DM for details🐘 📩
m.hanson@exeter.ac.uk
Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria
nature.com/articles/s41467-024…
#evolution #microbiology #genomics
Stochasticity, determinism, and contingency shape genome evolution of endosymbiotic bacteria - Nature Communications
Endosymbionts often have small genomes that maintain minimal functions required to serve their hosts. This study examines cases of new endosymbiont acquisition and finds genome degeneration involves both stochastic and deterministic processes that sh…Nature
1) Want to know how much of your metagenome is eukaryotic? No references? No problem. We developed SingleM microbial fraction (SMF) and ran it on 250k metagenomes biorxiv.org/content/10.1101/20….
If you know what Eukaryotes are there, you can filter reads by mapping to their genomes. However, often you don’t know what’s in your sample, or the euk doesn’t have a genome.
#metagenomics #bioinformatics #genomics #microbiomes #microbialecology

Large-scale estimation of bacterial and archaeal DNA prevalence in metagenomes reveals biome-specific patterns
Metagenomes often contain many reads derived from eukaryotes. However, there is usually no reliable method for estimating the prevalence of non-microbial reads in a metagenome, forcing many analysis techniques to make the often-faulty assumption that…bioRxiv
Widespread occurrence and diverse origins of polintoviruses influence lineage-specific genome dynamics in stony corals
academic.oup.com/ve/advance-ar…
#viruses #coral #evolution #genomics

Widespread occurrence and diverse origins of polintoviruses influence lineage-specific genome dynamics in stony corals
Abstract:. Stony corals (Order Scleractinia) are central to vital marine habitats known as coral reefs. Numerous stressors in the Anthropocene are contribuStephens, Danae (Oxford University Press)
Systematic identification of cargo-mobilizing genetic elements reveals new dimensions of eukaryotic diversity
Interested in evolutionary genomics/population genomics and plant genetics? Want to do your PhD in beautiful Stockholm?
We have two 4-year PhD student positions available in my group at Stockholm University. More info, see tanjaslottelab.se
Please repost.
#evolution #genomics #popgen #distyly #CropWildRelatives #PlantGenetics #ecrchat #phd 1/4
#Genomics #Epigenetics #Bioinformatics
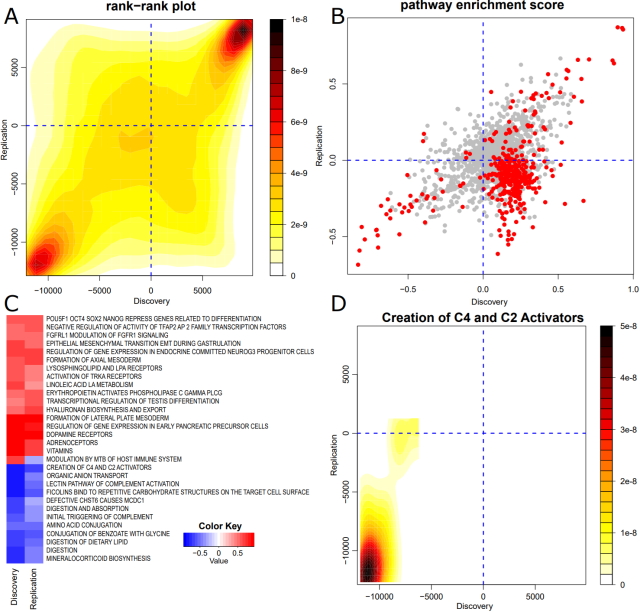
In this article we outline a refined method for pathway enrichment of infinium array data that is more sensitive and precise as compared to existing over-representation approaches. Feedback welcome.
biorxiv.org/content/10.1101/20…
Direction-aware functional class scoring enrichment analysis of Infinium DNA methylation data
Infinium Methylation BeadChip arrays remain one of the most popular platforms for epigenome-wide association studies, but tools for downstream pathway analysis have their limitations.bioRxiv
Large language models improve annotation of prokaryotic viral proteins
nature.com/articles/s41564-023…
#virology #viruses #bioinformatics #genomics
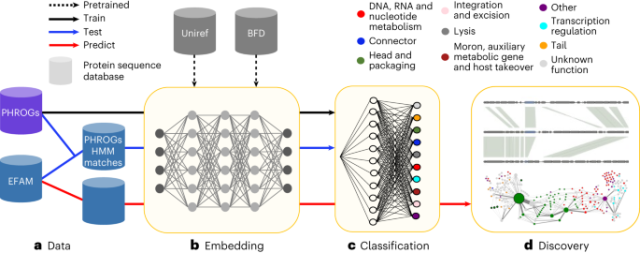
Large language models improve annotation of prokaryotic viral proteins - Nature Microbiology
Ocean viral proteome annotations are expanded by a machine learning approach that is not reliant on sequence homology and can annotate sequences not homologous to those seen in training.Nature
Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy
nature.com/articles/s41587-023…
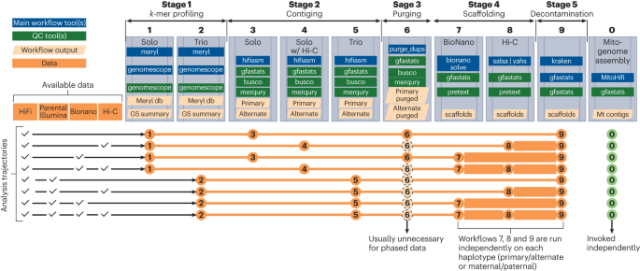
Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy - Nature Biotechnology
Nature Biotechnology - Scalable, accessible and reproducible reference genome assembly and evaluation in GalaxyNature
Gotta check these out!
Robust, scalable, and informative clustering for diverse biological networks
genomebiology.biomedcentral.co…
#bioinformatics #genomics #genetics #statistics

Robust, scalable, and informative clustering for diverse biological networks - Genome Biology
Clustering molecular data into informative groups is a primary step in extracting robust conclusions from big data.BioMed Central
Apply for our Phd project on "Discovering and characterising novel defence systems in pathogenic Serratia spp using microbiology and genomics"
#genomics #microbiology #omics
FindAPhD : Pathogens and Host Defences Doctoral Training Partnership PhD Studentships at University of Sussex
findaphd.com/phds/program/path…

FindAPhD : Pathogens and Host Defences Doctoral Training Partnership PhD Studentships at University of Sussex
Apply for a PhD: Pathogens and Host Defences Doctoral Training Partnership PhD Studentships at University of Sussexwww.FindAPhD.com
Gene duplication as a major force driving the genome expansion in some giant viruses
Ongoing shuffling of protein fragments diversifies core viral functions linked to interactions with bacterial hosts
nature.com/articles/s41467-023…
#phages #viruses #evolution #genomics
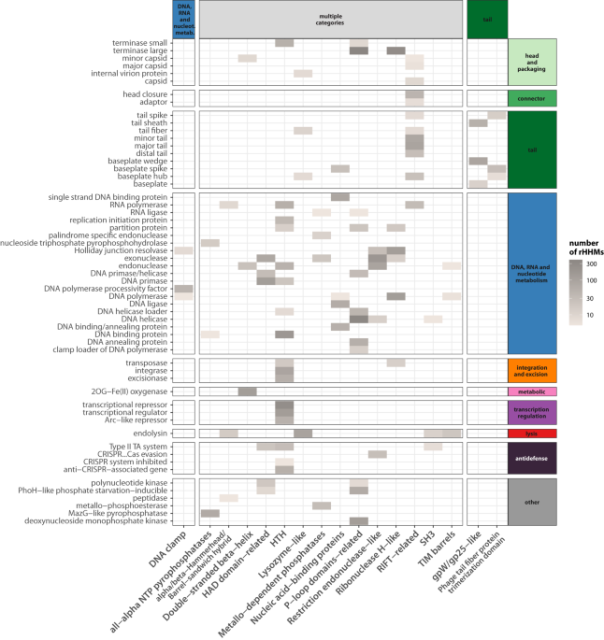
Ongoing shuffling of protein fragments diversifies core viral functions linked to interactions with bacterial hosts - Nature Communications
Proteins are composed of distinct functional domains, each serving a specific role. Here, Smug et al. show that phages are able to shuffle fragments of their proteins and this predominantly occurs in proteins involved in bacterial host interactions.Nature
Postdoctoral Fellow in Phylogenomics/Bioinformatics
#academicjobs #postdocjobs #genomics #evolution #microbiology
jobrxiv.org/job/uc-santa-barba…

Postdoctoral Fellow in Phylogenomics/Bioinformatics
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
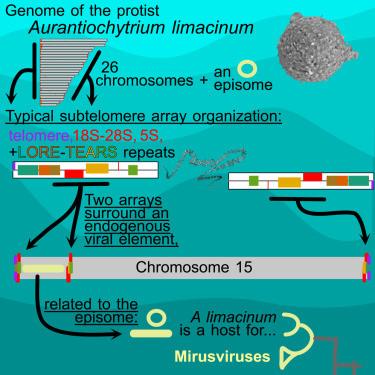
New #ISEPpapers! The #protist #Aurantiochytrium has universal subtelomeric rDNAs and is a host for #mirusviruses: Jackie Collier et al. cell.com/current-biology/fullt…
#protists #microbes #protistology #microbiology #viruses #virology #genomics
PhD position in viral evolution and diversity @foaylward
Virginia Tech
Funded PhD positions in the Aylward Lab to study the #evolution and #genomics of giant #viruses. Both computational and wet-lab projects available.
See the full job description on jobRxiv: jobrxiv.org/job/virginia-tech-…
#ScienceJobs #hiring #research
Blacksburg #UnitedStatesUS ...
jobrxiv.org/job/virginia-tech-…

PhD position in viral evolution and diversity
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
My latest preprint exploring transposable element (TE) activity in the genomes of sparrows.
biorxiv.org/cgi/content/short/…
We found remarkably high levels of repeat content in the newly generated genomes of
Bell's, Song, and Savannah Sparrow. 31% of the Bell's sparrow genome is spanned by repetitive elements.
This is ~3x as much as previously reported in most songbirds.
Thanks to all my co-authors for their contributions to this project!
#Genomics
#TransposableElements
#ornithology
#sparrows
Remarkably high repeat content in the genomes of sparrows: the importance of genome assembly completeness for transposable element discovery.
Transposable elements (TE) play critical roles in shaping genome evolution. However, the highly repetitive sequence content of TEs is a major source of assembly gaps.bioRxiv
Long-read-based genome assembly reveals numerous endogenous viral elements in the green algal bacterivore Cymbomonas tetramitiformis
Abstract. The marine tetraflagellate Cymbomonas tetramitiformis has drawn attention as an early diverging green alga that uses a phago-mixotrophic mode of nutriGyaltshen, Yangtsho (Oxford University Press)
PhylteR: efficient identification of outlier sequences in phylogenomic datasets.
PhylteR can automatically identify sequences likely to be hidden paralogs or horizontally transferred genes in very large datasets. Removing those sequences therefore reduces noise in downstream analyses.
Available as an R package on CRAN or as docker and singularity images.
Package:
cran.r-project.org/web/package…
Paper:
Ancestral genome reconstructions are changing the field of #comparative #genomics. Want to learn more?
Watch the talks by Hugues Roest Crollius & Matthieu Muffato in the last #ERGA BioGenome Analysis and Applications Seminar!
👉youtube.com/watch?v=9QDgRHlLdm…
Learn more about the seminar series & stay tuned for the upcoming sessions! 👉erga-biodiversity.eu/post/erga…
Earth BioGenome Project Biodiversity Genomics Europe

ERGA BioGenome Analysis and Applications Seminars
The ERGA BioGenome Analysis and Applications Seminars represent a joint endeavor of the ERGA Data Analysis Committee (DAC) and the Biodiversity Genomics Europe Work Package 11 - Genome Applications.erga
Bacterial histones unveiled
nature.com/articles/s41564-023…
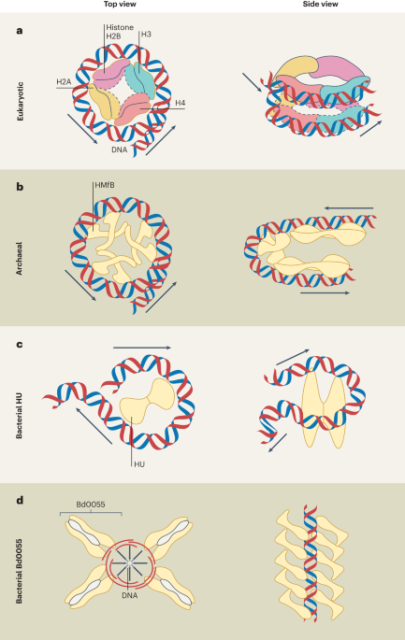
Bacterial histones unveiled - Nature Microbiology
Computational, molecular and structural analyses reveal the presence of bacterial histones that bind DNA to form dense, DNA-enveloping fibres in Bdellovibrio bacteriovorus.Nature
New Environmental #Bioinformatics Group
@SIB - #DataScience solutions to address environmental challenges sib.swiss/environmental-bioinf… 💻🧬📊🌍🛜🌦️🌄📋🔭

Robert M. Waterhouse
Waterhouse Group Arthropoda Assembly Assessment Catalogue | DrosOMA Orthology Database | GO-Figure! Gene Ontology Visualisationrmwaterhouse.org
'Both genomes had been analysed before, but the fragmented assemblies had scaffold sizes comparable to the length of long reads prior to assembly. Our new assemblies illustrate how long-read technologies allow for a much better representation of species genomes. We are now able to conduct more accurate downstream assays based on more complete gene and transposable element predictions.'
#Preprint #Evolution #Genomics
biorxiv.org/content/10.1101/20…

Revisiting genomes of non-model species with long reads yields new insights into their biology and evolution
High-quality genomes obtained using long-read data allow not only for a better understanding of heterozygosity levels, repeat content, and more accurate gene annotation, and prediction when compared to those obtained with short-read technologies, but…bioRxiv
#genomics #bioinformatics
biorxiv.org/content/10.1101/20…

Automated Bioinformatics Analysis via AutoBA
With the fast-growing and evolving omics data, the demand for streamlined and adaptable tools to handle the analysis continues to grow.bioRxiv
Cool paper! Also glad we are moving towards Patescibacteria and not CPR:
"Genetic manipulation of Patescibacteria provides mechanistic insights into microbial dark matter and the epibiotic lifestyle"
Preprint from Salzberg team questioning a 2020 Nature paper from Rob Knight 😮
"the raw read counts were vastly over-estimated for nearly every bacterial species, often by a factor of 1000 or more."
"Our conclusion after re-analysis is that the near-perfect association between microbes and cancer types reported in the study is, simply put, a fiction."
Major data analysis errors invalidate cancer microbiome findings
biorxiv.org/content/10.1101/20…
#microbiome #genomics #research #science

Major data analysis errors invalidate cancer microbiome findings
We re-analyzed the data from a recent large-scale study that reported strong correlations between microbial organisms and 33 different cancer types, and that created machine learning predictors with near-perfect accuracy at distinguishing among cance…bioRxiv
I wrote a short guide on how to build both alignment-free and reference-based prokaryote phylogenetic trees from SNP alignments without using snippy, check it out
bacpop.org/guides/building_tre…
Building trees with SKA
SKA is a tool for comparing small and highly similar genomes using split k-mers. This guide will explain how to use SKA to build a phylogenetic tree for different Escherichia coli lineages in a few minutes.www.bacpop.org
wur.nl/en/vacancy/phd-research…
#PhD #PhDPosition #Evolution #Genomics #ProteinStructure #MachineLearning