Search
Items tagged with: MicrobialEcology
Postdoctoral Fellow in Environmental Microbial Genomics
Trent University
Excited about microbial genomics, environmental change, and single-cell analysis? Join us as a postdoc @ecochangegroup!
See the full job description on jobRxiv: jobrxiv.org/job/trent-universi…
#microbialecology #microbiology #mi...
jobrxiv.org/job/trent-universi…

Postdoctoral Fellow in Environmental Microbial Genomics
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
An interesting #PlantSciJob as Editor at Nature Microbiology @natmicrobiol.nature.com. Expertise in #PlantMicrobiology #MicrobialEcology #PlantImmunity desirable. #PlantScience #PlantSciJobs #PlantScienceJobs
1) Want to know how much of your metagenome is eukaryotic? No references? No problem. We developed SingleM microbial fraction (SMF) and ran it on 250k metagenomes biorxiv.org/content/10.1101/20….
If you know what Eukaryotes are there, you can filter reads by mapping to their genomes. However, often you don’t know what’s in your sample, or the euk doesn’t have a genome.
#metagenomics #bioinformatics #genomics #microbiomes #microbialecology

Large-scale estimation of bacterial and archaeal DNA prevalence in metagenomes reveals biome-specific patterns
Metagenomes often contain many reads derived from eukaryotes. However, there is usually no reliable method for estimating the prevalence of non-microbial reads in a metagenome, forcing many analysis techniques to make the often-faulty assumption that…bioRxiv
Happy to see the first experimental project I led finally out in the world! With Jeremy Moore, Sydney Olsen, and Mike Travisano
We combined experimental evolution and mathematical modeling to ask: do bacteria evolve to swim away from phages?
1/n
doi.org/10.1101/2023.04.29.538…
#microbiology #evolution #ecology #phages #phage #bacteria #MicrobialEvolution #MicrobialEcology

Fight not flight: parasites drive the bacterial evolution of resistance, not avoidance
In the face of ubiquitous threats from parasites, hosts often evolve strategies to resist infection or to altogether avoid contact with parasites. At the microbial scale, bacteria frequently encounter viral parasites, bacteriophages.bioRxiv
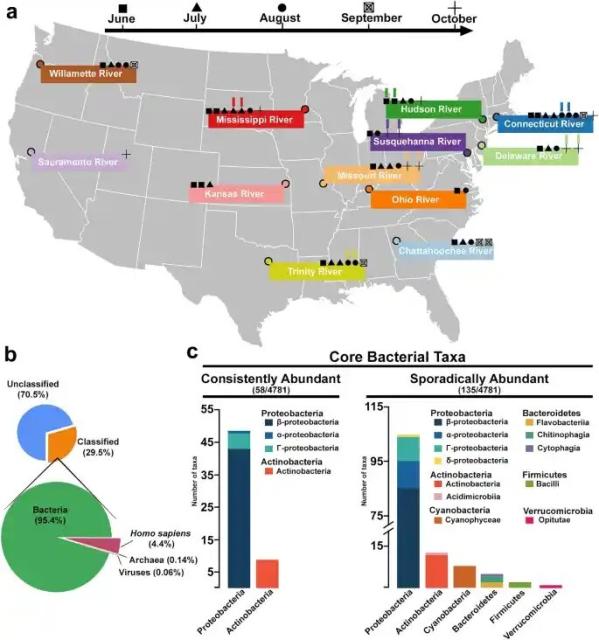
Blooms of #Cyanobacteria pose a significant threat to #freshwater systems including rivers driving both #eutrophication and its dire implications for freshwater species as oxygen availability plummets and also the additional threat posed by #cyanotoxins produced by some Cyanobacteria.
This new research in Scientific Reports takes a #metagenomics and Q-PCR approach to explore the composition of the Cyanobacterial populations and their cyanotoxin production gene in major rivers across the United States (in 2019) and identifies Microcystis as the key toxin producing genus across this study.
Find out more on:
nature.com/articles/s41598-023…
#microbiology #ecology #MicrobialEcology
Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United States - Scientific Reports
Scientific Reports - Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United StatesNature
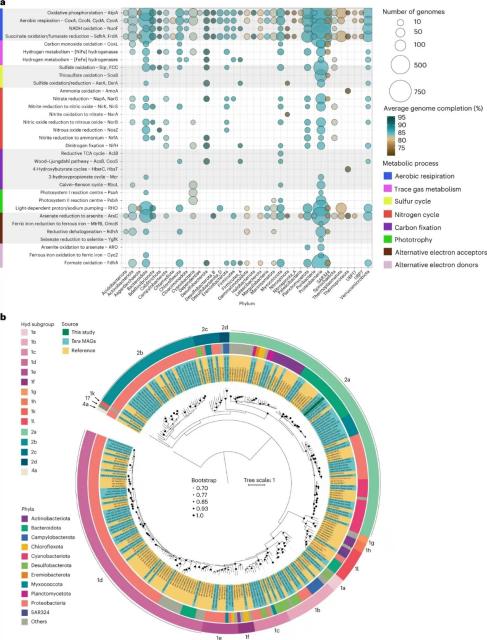
As we (humans) get to grips with how to scale up #hydrogen as an energy source (and for energy storage) and as part replacement for #fossilfuels, this new research (once again) shows that #microbes have beaten us to it and evolved their own solutions for energy - in this case using hydrogen power.
This new research in Nature Microbiology explores how hydrogen in seawater supports growth of multiple and diverse #bacterial species.
Fof those (like me) in #Melbourne , you may be interested that the research also includes study of this process within #PortPhillipBay sediments.
Learn more on:
nature.com/articles/s41564-023…
#microbiology #bacteria #ecology #MicrobialEcology
Molecular hydrogen in seawater supports growth of diverse marine bacteria - Nature Microbiology
Genome-resolved metagenomics, biogeochemistry, modelling and culture-based analysis reveal that marine bacteria consume H2 to support growth.Nature
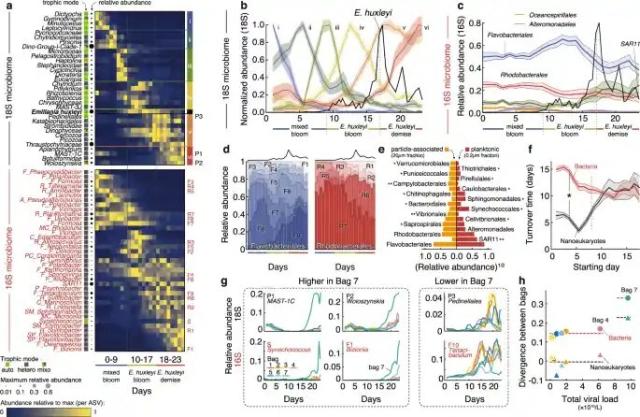
#Virus control
This exciting new research explores the influence of #virus #infection in changing the balance of organic matter degradation between #bacteria and #eukaryotes in #marine #carboncycling during #phytoplankton blooms.
Based on #mesocosm studies, this research demonstrates how #viruses infecting different #microorganisms change community dynamics and the release and potential fate of marine carbon from primary production.
Learn more in this new research in Nature Communications on:
nature.com/articles/s41467-023…
#microbiology #ecology #MicrobialEcology
Viral infection switches the balance between bacterial and eukaryotic recyclers of organic matter during coccolithophore blooms - Nature Communications
Algal blooms are hotspots of marine primary production that play central roles in microbial ecology and global elemental cycling.Nature