Search
Items tagged with: metagenomics
1) Want to know how much of your metagenome is eukaryotic? No references? No problem. We developed SingleM microbial fraction (SMF) and ran it on 250k metagenomes biorxiv.org/content/10.1101/20….
If you know what Eukaryotes are there, you can filter reads by mapping to their genomes. However, often you don’t know what’s in your sample, or the euk doesn’t have a genome.
#metagenomics #bioinformatics #genomics #microbiomes #microbialecology

Large-scale estimation of bacterial and archaeal DNA prevalence in metagenomes reveals biome-specific patterns
Metagenomes often contain many reads derived from eukaryotes. However, there is usually no reliable method for estimating the prevalence of non-microbial reads in a metagenome, forcing many analysis techniques to make the often-faulty assumption that…bioRxiv
Metabarcoding, metagenomics and other omics methods revolutionize the study and monitoring of aquatic ecosystems. Come and join the 350 scientists and managers who will be discussing the use and development of these methods.
aquaecomics.symposium.inrae.fr…
#metabarcoding #metagenomics #fish #algae #protist #DNA #bioinformatic #pipeline #omics #MolecularEcology

Postdoc position in microbial OMICS and microplastics (fixed-term for 36 months) (m/f/x) at Leibniz-Institut für Gewässerökologie und Binnenfischerei
We're looking for: Postdoc position in microbial OMICS and microplastics (fixed-term for 36 months) (m/f/x) (Full or Part Time) • Neuglobsow, Stechlin, Deutschlandkarriere-igb.softgarden.io
A genome catalogue of lake bacterial diversity and its drivers at continental scale
nature.com/articles/s41564-023…
#metagenomics #lakes #freshwater #microbiology
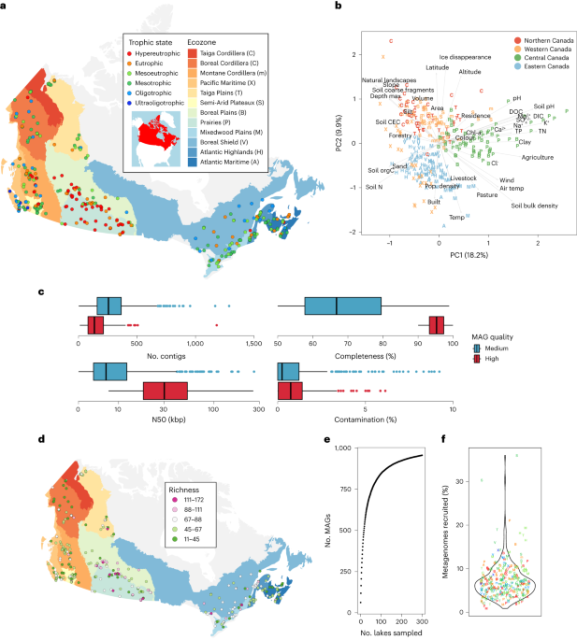
A genome catalogue of lake bacterial diversity and its drivers at continental scale - Nature Microbiology
A metagenomic compendium from lakes across Canada reveals bacterial metabolic potential and how it varies depending on land use in different regions.Nature
Characterization and simulation of metagenomic nanopore sequencing data with Meta-NanoSim
academic.oup.com/gigascience/a…

Characterization and simulation of metagenomic nanopore sequencing data with Meta-NanoSim
AbstractBackground. Nanopore sequencing is crucial to metagenomic studies as its kilobase-long reads can contribute to resolving genomic structural differencesYang, Chen (Oxford University Press)
VirBot: an RNA viral contig detector for metagenomic data
academic.oup.com/bioinformatic…

VirBot: an RNA viral contig detector for metagenomic data
AbstractSummary. Without relying on cultivation, metagenomic sequencing greatly accelerated the novel RNA virus detection. However, it is not trivial to accuratChen, Guowei (Oxford University Press)
TAMPA: interpretable analysis and visualization of metagenomics-based taxon abundance profiles
academic.oup.com/gigascience/a…

TAMPA: interpretable analysis and visualization of metagenomics-based taxon abundance profiles
AbstractBackground. Metagenomic taxonomic profiling aims to predict the identity and relative abundance of taxa in a given whole-genome sequencing metagenomic sSarwal, Varuni (Oxford University Press)
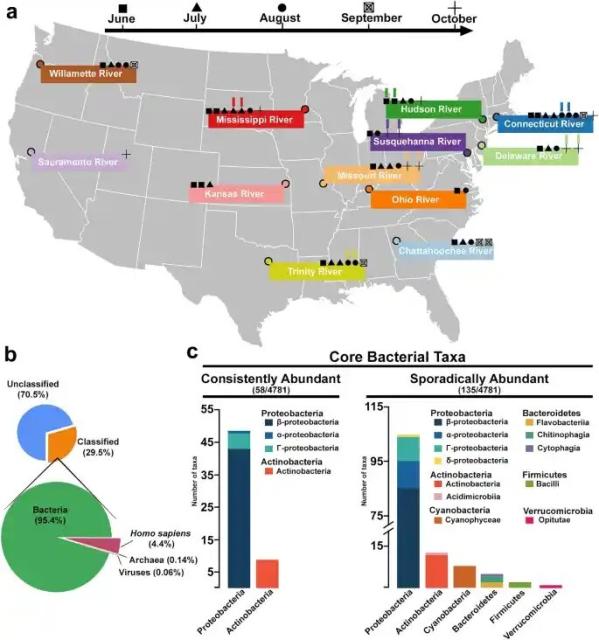
Blooms of #Cyanobacteria pose a significant threat to #freshwater systems including rivers driving both #eutrophication and its dire implications for freshwater species as oxygen availability plummets and also the additional threat posed by #cyanotoxins produced by some Cyanobacteria.
This new research in Scientific Reports takes a #metagenomics and Q-PCR approach to explore the composition of the Cyanobacterial populations and their cyanotoxin production gene in major rivers across the United States (in 2019) and identifies Microcystis as the key toxin producing genus across this study.
Find out more on:
nature.com/articles/s41598-023…
#microbiology #ecology #MicrobialEcology
Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United States - Scientific Reports
Scientific Reports - Metagenomic mapping of cyanobacteria and potential cyanotoxin producing taxa in large rivers of the United StatesNature
We are all too familiar with the all-too-often catastrophic impacts of #COVID19 #infection on human #health.
This fascinating new #metagenomics research in mBio explores a potentially important and intriguing link between the presence and relative abundance of different functional guilds present in the human gut #microbiome (including short chain fatty acid biosynthesis and virulence functions) and disease severity and outcomes resulting from the virus. Clearly a subject for future extended studies to explore potential increases in severity due to infection and exacerbated or mitigated by differences in gut microbiomes.
Explore the research on: