Search
Items tagged with: bioinformatics
Heng Li: hlilab.github.io/vacancies
"We are looking for a postdoctoral fellow to develop high-performance algorithms for sequence alignment, pangenome representation, application of pangenome data structures, or other projects the applicant may propose. A qualified candidate is expected to be proficient in a high-performance language such as C, C++ or Rust, and have rich research experiences in #bioinformatics and computational genomics."
#job
#bioinformatics
bioinformatics.ca/workshops-al…
#bioinformatics
tinyurl.com/3aewk286
#EvoDevo #MolecularEvolution #bioinformatics #PopulationGenomics #RNAseq #PostDoc
Excited to announce our article, "EvoWeaver: large-scale prediction of gene functional associations from coevolutionary signals" was just published in Nature Communications!
We demonstrate that it's possible to infer how genes work together using only sequencing data by leveraging correlated signals of evolution. Check it out at the link below!
#genomics #bioinformatics #rstats #genetics #evolution #biology
nature.com/articles/s41467-025…
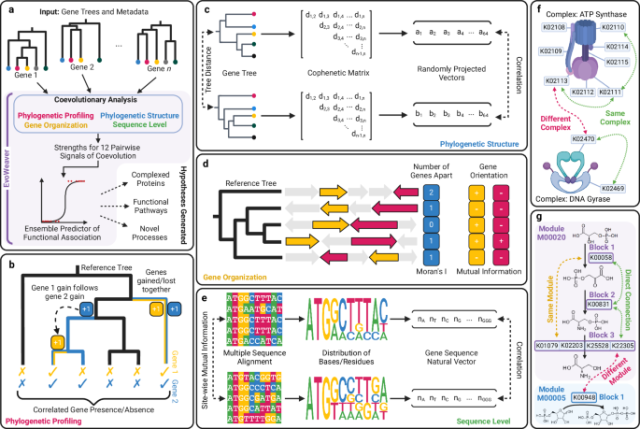
EvoWeaver: large-scale prediction of gene functional associations from coevolutionary signals - Nature Communications
Despite having structures for most proteins, we still do not know their function. Here, authors introduce EvoWeaver, a de novo method to identify genes working together using evolutionary information.Nature
Are you getting into the #bioinformatics area coming from a #biological sciences background?
This new blog post is for you!
How To Operate Large Files in Bioinformatics
Definitely my past self would have benefited from it. Enjoy!
What do #nanopore #sequencing #bioinformatics folks think about this one? New preprint on hashing raw nanopore signals and using them directly for de-novo assembly.
Hypothetical blow5-to-assembly pipe would be interesting, though figuring it out is way out of my depth.

Enabling Fast, Accurate, and Efficient Real-Time Genome Analysis via New Algorithms and Techniques
The advent of high-throughput sequencing technologies has revolutionized genome analysis by enabling the rapid and cost-effective sequencing of large genomes.arXiv.org
New shitty reality:
Every time #NCBI is #down you wonder whether it's permanent or not.
Just now the whole nih.gov is not resolving from two different locations I have access to.
Also:
downdetector.com/status/nih/
And just in time when I decided to do some #bioinformatics work 1AM on Sat^W Sunday.
#nih
I didn't know PacBio maintained its own curation of sample/reference datasets. Saves me a bit of trouble of hunting down reliable ones for a pipeline testing:

Datasets - PacBio - Highly accurate long-read sequencing
Explore these publicly available datasets generated using SMRT Sequencing applications including whole genome sequencing, RNA sequencing, and more.PacBio
Modeling the mosaic structure of bacterial genomes to infer their evolutionary history
#evolution #genomics #bacteria #bioinformatics #science
New tool from me: I wrote a local aligner that can do approximate reference-based alignment and find genes in a query microbial genome, without a separate indexing step or any temporary disk space usage.
Written fully in Rust and there will be a browser version out later: github.com/tmaklin/kbo 1/3
#rust #rustlang #bioinformatics
GitHub - tmaklin/kbo: Spectral Burrows-Wheeler transform accelerated local alignment search.
Spectral Burrows-Wheeler transform accelerated local alignment search. - tmaklin/kboGitHub
ORA is a type of enrichment analysis that analyses over-represented functional categories in gene lists. These tools have accumulated ~190k citations, but they have subtly different behaviours. Here we unpack the differences and investigate two subtle problems in some implementations, which may have negatively impacted those 190k research papers.
doi.org/10.1093/bioadv/vbae159
#genomics #bioinformatics
Our new preprint on the impact of protein phosphorylation on structures is now out!
Protein phosphorylation is a key regulator of cellular processes.It's ubiquitous, yet the function and relevance of most phosphosites is unknown. Understanding phosphosite function requires elucidating the structural mechanisms through which it acts. (1)
biorxiv.org/content/10.1101/20…
@bioinformatics @strucbio #bioinformatics #protein #StructuralBiology

Global comparative structural analysis of responses to protein phosphorylation
Post-translational modifications (PTMs), particularly protein phosphorylation, are key regulators of cellular processes, impacting numerous aspects of protein activity.bioRxiv
Profile and Binomica Labs registered on archaea.bio!
I'll be doing a lot more Halobacteria & Halococcus work in the near future, looking forward to sharing some interesting notes
archaea.bio/profiles/sung-won-…
#microbiology #archaea #bioinformatics

Sung won Lim
Independent researcher at Binomica Labs (Queens, NYC), and a remote adjunct researcher in Kyle lab at University of New Hampshire.Sung won Lim (archaea.bio)
The SplitsTree App: interactive analysis and visualization using phylogenetic trees and networks
nature.com/articles/s41592-024…
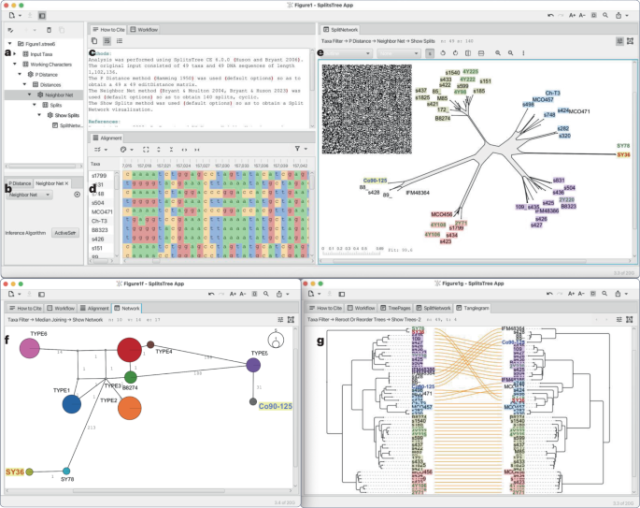
The SplitsTree App: interactive analysis and visualization using phylogenetic trees and networks - Nature Methods
Nature Methods - The SplitsTree App: interactive analysis and visualization using phylogenetic trees and networksNature
PhD Student in Virology/Bionformatics
Otto von Guericke Universität Magdeburg
The project will investigate genomic features in DVG and DI formation in #enteroviruses using #molecular biology and #bioinformatics.
See the full job description on jobRxiv: jobrxiv.org/job/otto-von-gueri…...
jobrxiv.org/job/otto-von-gueri…

PhD Student in Virology/Bionformatics
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
Funded PhD Position in metagenomics of environmental films
Texas Tech University
Exciting funded PhD opportunity studying metagenomics of environmental films
See the full job description on jobRxiv: jobrxiv.org/job/texas-tech-uni…
#biofilms #bioinformatics #biology #metag...
jobrxiv.org/job/texas-tech-uni…
Funded PhD Position in metagenomics of environmental films
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
1) Want to know how much of your metagenome is eukaryotic? No references? No problem. We developed SingleM microbial fraction (SMF) and ran it on 250k metagenomes biorxiv.org/content/10.1101/20….
If you know what Eukaryotes are there, you can filter reads by mapping to their genomes. However, often you don’t know what’s in your sample, or the euk doesn’t have a genome.
#metagenomics #bioinformatics #genomics #microbiomes #microbialecology

Large-scale estimation of bacterial and archaeal DNA prevalence in metagenomes reveals biome-specific patterns
Metagenomes often contain many reads derived from eukaryotes. However, there is usually no reliable method for estimating the prevalence of non-microbial reads in a metagenome, forcing many analysis techniques to make the often-faulty assumption that…bioRxiv
"Indexing All Life's Known Biological #Sequences" biorxiv.org/content/10.1101/20…
"In this work, we take advantage of recently developed, very efficient data structures and algorithms for representing sequence sets. We make Petabases of DNA sequences across all clades of life, including viruses, bacteria, fungi, plants, animals, and humans, fully searchable and make the indexes available to the research community"

Indexing All Life's Known Biological Sequences
The amount of biological sequencing data available in public repositories is growing exponentially, forming an invaluable biomedical research resource.bioRxiv
Hello sugar people of the  " title="
" title="![]() "/> . My former collegue Bernard Henrissat now in 🇩🇰 is looking for a #PhD student in a Marie Sklodowska-Curie 🇪🇺 training network to work with him at the Technical University of Denmark. More info:
"/> . My former collegue Bernard Henrissat now in 🇩🇰 is looking for a #PhD student in a Marie Sklodowska-Curie 🇪🇺 training network to work with him at the Technical University of Denmark. More info:
euraxess.ec.europa.eu/jobs/201…
The deadline for application is May 31st, 2024 and the job will start in November.
Skills desired: #bioinformatics, general biology (++ #carbohydrates) and fluency in 🇬🇧.
Eligibility: the candidate should not have worked in 🇩🇰 before.
Conda is notorious slow... then I found mamba github.com/mamba-org/mamba, then I found
pixi prefix.dev/blog/pixi_a_fast_co… : Blazing fast cross-platform package management for teams.
By the creators of the mamba package manager. #bioinformatics
GitHub - mamba-org/mamba: The Fast Cross-Platform Package Manager
The Fast Cross-Platform Package Manager. Contribute to mamba-org/mamba development by creating an account on GitHub.GitHub
There's a new minimap2 release with ONT specific accurate long reads setting. Looking forward to giving it a test run!
github.com/lh3/minimap2/releas…

Release Minimap2-2.27 (r1193) · lh3/minimap2
Notable changes to minimap2: New feature: added the lr:hq preset for accurate long reads at ~1% error rate. This was suggested by Oxford Nanopore developers (#1127). It is not clear if this prese...GitHub
#Genomics #Epigenetics #Bioinformatics
In this article we outline a refined method for pathway enrichment of infinium array data that is more sensitive and precise as compared to existing over-representation approaches. Feedback welcome.
biorxiv.org/content/10.1101/20…
Direction-aware functional class scoring enrichment analysis of Infinium DNA methylation data
Infinium Methylation BeadChip arrays remain one of the most popular platforms for epigenome-wide association studies, but tools for downstream pathway analysis have their limitations.bioRxiv
Looking for some suggestions - I'm working with Archaeal genomic regions whose optimally fitting model seems to be Tamura-Nei (doi.org/10.1093/oxfordjournals…).
Would it be too much to suspect the Archaeal region is under similar types of evolutionary pressure?
#archaea #microbiology #evolution #phylogenetics #bioinformatics
Large language models improve annotation of prokaryotic viral proteins
nature.com/articles/s41564-023…
#virology #viruses #bioinformatics #genomics
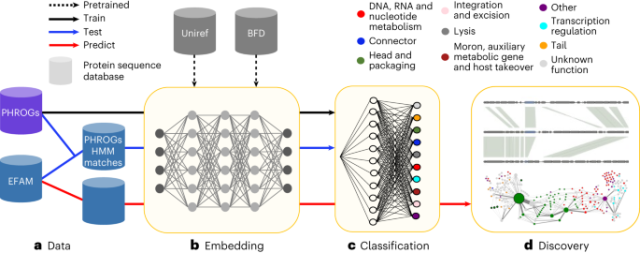
Large language models improve annotation of prokaryotic viral proteins - Nature Microbiology
Ocean viral proteome annotations are expanded by a machine learning approach that is not reliant on sequence homology and can annotate sequences not homologous to those seen in training.Nature
Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy
nature.com/articles/s41587-023…
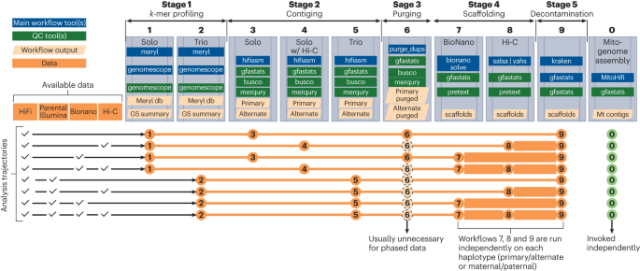
Scalable, accessible and reproducible reference genome assembly and evaluation in Galaxy - Nature Biotechnology
Nature Biotechnology - Scalable, accessible and reproducible reference genome assembly and evaluation in GalaxyNature
Gotta check these out!
Robust, scalable, and informative clustering for diverse biological networks
genomebiology.biomedcentral.co…
#bioinformatics #genomics #genetics #statistics

Robust, scalable, and informative clustering for diverse biological networks - Genome Biology
Clustering molecular data into informative groups is a primary step in extracting robust conclusions from big data.BioMed Central
Stowers is starting up a paid training program for bioinformatics, please share with anyone who might be interested in something like that! (in Kansas City)
stowers.org/gradschool/computa…

Computational Biology Scholars
The mission of The Graduate School of the Stowers Institute for Medical Research is to prepare a superb cadre of predoctoral researchers from around the world for the pursuit of innovative and creative investigations in the biological sciences.Turnstyle
doi.org/10.1038/s43705-023-003…
#microbiology #bioinformatics
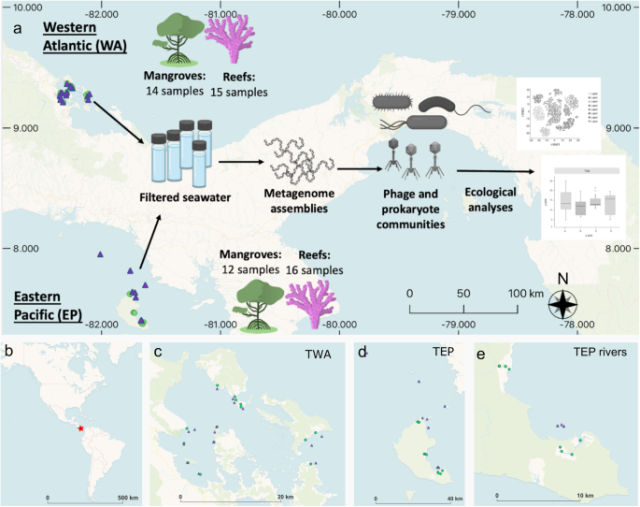
Contrasting drivers of abundant phage and prokaryotic communities revealed in diverse coastal ecosystems - ISME Communications
ISME Communications - Contrasting drivers of abundant phage and prokaryotic communities revealed in diverse coastal ecosystemsNature
If anyone is looking for the WGCNA tutorials while the Horvath's lab website has been down for a while, there's a Dropbox link with all the tutorials here: bioinformatics.stackexchange.c…

Where to access the WGCNA tutorial documents: Horvath lab site down
I am currently using the WGCNA package for some analysis and it seems the Horvath lab site is down. Does anyone know of anywhere else I can access the tutorial documents?Bioinformatics Stack Exchange
Database resources of the National Center for Biotechnology Information
#ReferenceSequence #Databases #NCBI #GenBank #PubMed #SRA #RefSeq #Taxonomy #bioinformatics
At my workplace, we're looking into how we can support the processing of very large datasets in R. It would be wonderful if some bioinformaticians could answer a couple of questions to direct us to the problem points.
We're hoping that we can publish something out of this that will be helpful to everyone in the field: forms.office.com/r/jNd2cbEZkh
OBF » OBF and BOSC leaving Twitter/X » OBF and BOSC leaving Twitter/X
Open Bioinformatics Foundation Homepagewww.open-bio.org
Come work with us! I have a PhD position available to help understand which mutations in the non-coding parts of coding transcripts have functional consequences in Stem Cells.
We will using genome wide mesaurement of the effect of variation in miRNA binding sites to address when vairation causes changes in regulation and when it doesn't.
Mixed wetlab, bioinformatics and statistics project.
findaphd.com/phds/project/whit…
#phdPosition #PhD #bioinformatics #UTR #miRNA #genetics

White Rose BBSRC DTP: How can we identify variants in untranslated RNA that affect stem-cells at University of Sheffield on FindAPhD.com
PhD Project - White Rose BBSRC DTP: How can we identify variants in untranslated RNA that affect stem-cells at University of Sheffield, listed on FindAPhD.comwww.FindAPhD.com
Very happy to share our new publication in PLOS ONE:
KIPEs3: Automatic annotation of biosynthesis pathways
doi.org/10.1371/journal.pone.0…
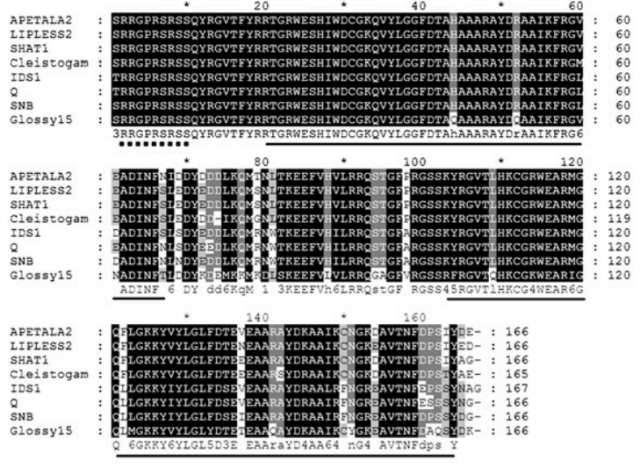
Excellent work by Andreas Rempel (Bielefeld University) and Nancy Choudhary (@PuckerLab @tubraunschweig #Bioinformatics #OpenAccess
KIPEs3: Automatic annotation of biosynthesis pathways
Flavonoids and carotenoids are pigments involved in stress mitigation and numerous other processes. Both pigment classes can contribute to flower and fruit coloration.doi.org
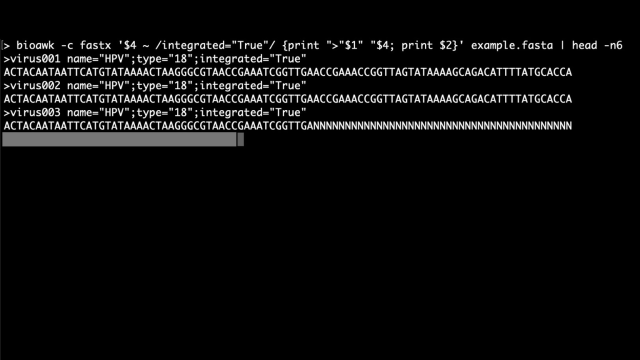
#bioawk is a command-line gem; it’s an extension of awk that auto-assigns variables for BED, SAM, VCF, GFF, and FASTX[AQ] format files, speeding up routine tasks.
For FASTX:
$1:name
$2:seq
$3:qual (FASTQ only)
$4:comment
Found in @vsbuffalo’s great #Bioinformatics Data Skills.
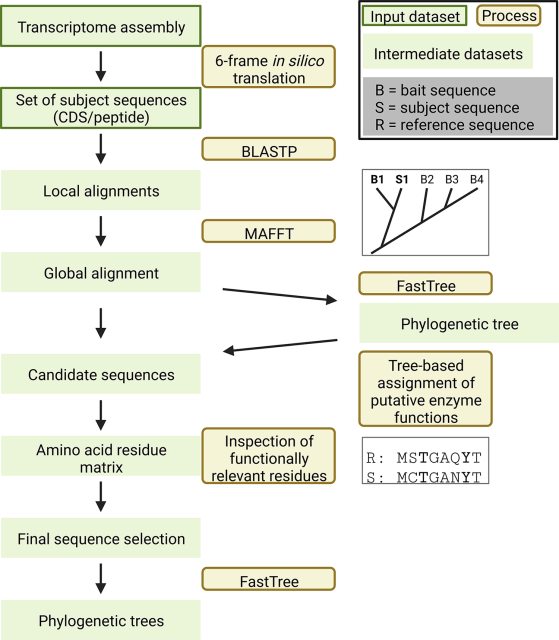
PhylteR: efficient identification of outlier sequences in phylogenomic datasets.
PhylteR can automatically identify sequences likely to be hidden paralogs or horizontally transferred genes in very large datasets. Removing those sequences therefore reduces noise in downstream analyses.
Available as an R package on CRAN or as docker and singularity images.
Package:
cran.r-project.org/web/package…
Paper: