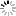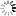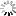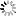Search
Items tagged with: phylogeny
A taxon-rich and genome-scale phylogeny of Opisthokonta
Opisthokonta is the major lineage that includes animals, fungi, and their unicellular relatives, but some ancient divergences remain contentious.plos.io
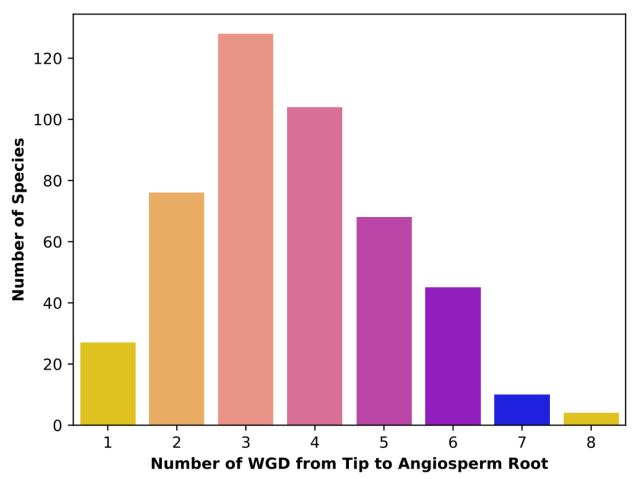
A new preprint from the lab on the distribution of ancient whole-genome duplications across the angiosperm phylogeny. Great work led by PhD student Michael McKibben! We used a variety of methods and different species trees to infer and place WGDs across the phylogeny. Overall, similar results to our past work, but species tree had a large impact on WGD inferences. Check it out here:
biorxiv.org/content/10.1101/20…
#WGD #polyploidy #phylogeny #angiosperms
Species Tree Topology Impacts the Inference of Ancient Whole-Genome Duplications Across the Angiosperm Phylogeny
Premise The history of angiosperms is marked by repeated rounds of ancient whole-genome duplications (WGDs).bioRxiv
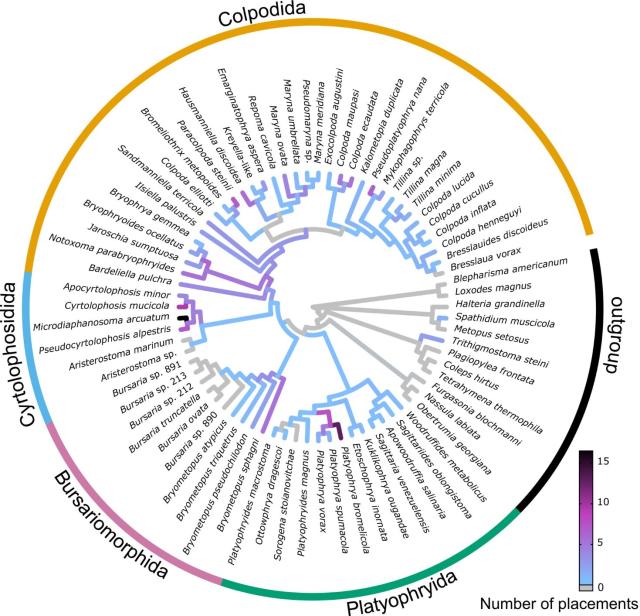
PhylteR: efficient identification of outlier sequences in phylogenomic datasets.
PhylteR can automatically identify sequences likely to be hidden paralogs or horizontally transferred genes in very large datasets. Removing those sequences therefore reduces noise in downstream analyses.
Available as an R package on CRAN or as docker and singularity images.
Package:
cran.r-project.org/web/package…
Paper:
biorxiv.org/content/10.1101/20…
#phylogeny #phylogenomics #bioinformatics #BigData #visualization #vizbi #myxozoan @dee_unil
1/thread
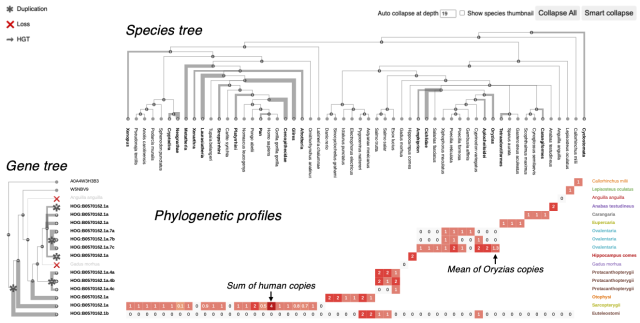
Matreex: compact and interactive visualisation of large gene families provides evidence for loss of intraflagellar transport in a myxozoan
Studying gene family evolution strongly benefits from insightful visualisations. However, the ever-growing number of sequenced genomes is leading to increasingly larger gene families, which challenges existing gene tree visualisations.bioRxiv