Search
Items tagged with: Bioinformatics
#PhD positions available in my group for Fall 2024 - projects focusing on #GiantVirus diversity and #evolution. Opportunities for both molecular wet-lab and #bioinformatics research.
The lab is fun, supportive, and inclusive, and we have many cool new viruses we are studying, so come join us!
Please boost and spread the word!
jobrxiv.org/job/virginia-tech-…

PhD position in viral evolution and diversity
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
insider.microsoft365.com/en-us…
#excel #dataanalysis #research #bioinformatics
Control data conversions in Excel for Windows and Mac
Based on your feedback, we've improved the Automatic Data Conversion settings, and made them also available in Excel for Mac.https://insider.office.com
Registrations are now open for the 2nd edition of the course "Analysis of Prokaryotic Pangenomes" with @jomcinerney & Alan Beavan.
Check it out: physalia-courses.org/courses-w…
Analysis of Prokaryotic Pangenomes
ONLINE, 15-17 April 2024 To foster international participation, this course will be held onlinephysalia-courses

Doctoral student in in structural and functional protein bioinformatics
Description of the workplace In the Atkinson lab we are interested in making discoveries about protein function and structure, with a focus on bacterial immune system components that protect againstlu.varbi.com
New Environmental #Bioinformatics Group
@SIB - #DataScience solutions to address environmental challenges sib.swiss/environmental-bioinf… 💻🧬📊🌍🛜🌦️🌄📋🔭

Robert M. Waterhouse
Waterhouse Group Arthropoda Assembly Assessment Catalogue | DrosOMA Orthology Database | GO-Figure! Gene Ontology Visualisationrmwaterhouse.org
#genomics #bioinformatics
biorxiv.org/content/10.1101/20…

Automated Bioinformatics Analysis via AutoBA
With the fast-growing and evolving omics data, the demand for streamlined and adaptable tools to handle the analysis continues to grow.bioRxiv
GitHub - arvestad/alv: A console-based alignment viewer
A console-based alignment viewer. Contribute to arvestad/alv development by creating an account on GitHub.GitHub
Anyone have advice on how to get the Spades genome assembler to use less memory?
GitHub - ablab/spades: SPAdes Genome Assembler
SPAdes Genome Assembler. Contribute to ablab/spades development by creating an account on GitHub.GitHub
I wrote a short guide on how to build both alignment-free and reference-based prokaryote phylogenetic trees from SNP alignments without using snippy, check it out
bacpop.org/guides/building_tre…
Building trees with SKA
SKA is a tool for comparing small and highly similar genomes using split k-mers. This guide will explain how to use SKA to build a phylogenetic tree for different Escherichia coli lineages in a few minutes.www.bacpop.org
After months of work I'm so happy that our new preprint is online!
Profiling the expression of #transportome genes in #cancer: a systematic approach.
biorxiv.org/content/10.1101/20…
It's a bit unfinished but it's by design! We want and need #feedback from #bioinformatics and #physiology people to move forward. I've tried to follow #openscience principles, so the pipeline is completely autonomous, containerized and the code for the paper is there, included in the repos!

Profiling the Expression of Transportome Genes in cancer: A systematic approach
The transportome, the -omic layer encompassing all Ion Channels and Transporters (ICTs), is crucial for cell physiology. It is therefore reasonable to hypothesize a role of the transportome in disease, and in particular in cancer.bioRxiv
I wanted to share with you a new thing I'm working on: biohandbook.me/
It's a resource for practical best practices and tips to use in bioinformatic analyses. The idea is to work together to define standards and workflows and write them down. I love the open science, FAIR data and FAIR code philosophy but there are not many easy-to-use and direct resources around to actually tell you how to do things in the bioinformatics world.
1/n
#bioinformatics #handbook #OpenScience
The Handbook
The Handbook # Welcome to Hedmad’s Handbook of Nice and Practical Tips! 🎉 This handbook aims to: Collect useful and practical day-to-day tips on how to conduct bioinformatics analyses; Set standards for data formats, workflows, project structure and …biohandbook.me
So many good points in this piece by
@PhilippBayer
My highlight in this screenshot (#science delivery & success is a TEAM effort!), but if you're vaguely involved in #academia +/- #bioinformatics, do have a read and think how it all may apply to you. #academicchatter @academicchatter
genomic.social/@PhilippBayer/1…
Hello everyone!
I’m an associate prof at Virginia Tech studying viral diversity. Lately my group has been focusing on giant viruses, but we are also interested in other protist viruses, and the role of viral endogenization in host genome evolution.
#GiantViruses #ViralDiversity #TreeOfLife #Protists #Microbiology #Bioinformatics #Evolution #Virology #Genomics
Recently we have been examining the strange and complex genes encoded in giant virus genomes!
doi.org/10.1093/femsre/fuad053
Studying some phage genes with scattered, partial alignments in varying degrees (40~+90%) of ID against closely related cousins (~93% ANI).
I'm thinking intragenic insertion/shuffling - but what kind of signals could I look for here? Common scars? Models to reference? #phage #bioinformatics #genomics
biorxiv.org/content/10.1101/20… #bioinformatics #HumanGenome #genomics

miniBUSCO: a faster and more accurate reimplementation of BUSCO
Motivation Assembly completeness evaluation of genome assembly is a critical assessment of the accuracy and reliability of genomic data. An incomplete assembly can lead to errors in gene predictions, annotation, and other downstream analyses.bioRxiv
Accounting for 16S rRNA copy number prediction uncertainty and its implications in bacterial diversity analyses
nature.com/articles/s43705-023…
#microbiology #bioinformatics #microbiome
Accounting for 16S rRNA copy number prediction uncertainty and its implications in bacterial diversity analyses - ISME Communications
ISME Communications - Accounting for 16S rRNA copy number prediction uncertainty and its implications in bacterial diversity analysesNature
doi.org/10.1038/s43705-023-002…
#microbiology #bioinformatics
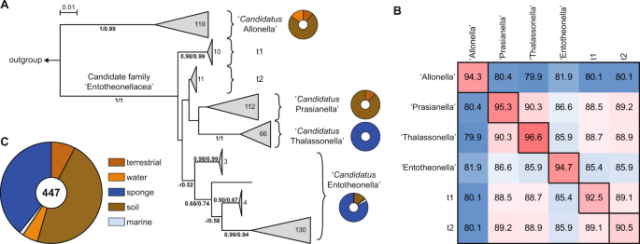
Distribution and diversity of ‘Tectomicrobia’, a deep-branching uncultivated bacterial lineage harboring rich producers of bioactive metabolites - ISME Communications
ISME Communications - Distribution and diversity of ‘Tectomicrobia’, a deep-branching uncultivated bacterial lineage harboring rich producers of bioactive metabolitesNature
Master or PhD positions
University of Ottawa
Master and PhD in #Canada fully funded positions in #bioinformatics
See the full job description on jobRxiv: jobrxiv.org/job/university-of-…
#ScienceJobs #hiring #research #bioinformatics #machine learning
Ottawa #Canada #PhDStudent #ResearchAssistant
jobrxiv.org/job/university-of-…

Master or PhD positions
Post a job in 3min, or find thousands of job offers like this one at jobRxiv!jobRxiv
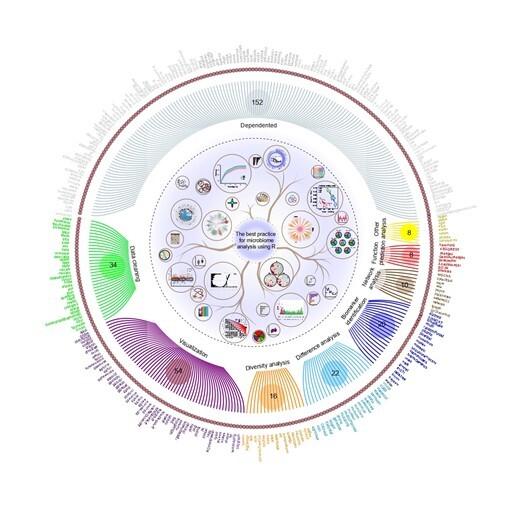
The best practice for microbiome analysis using R
Abstract. With the gradual maturity of sequencing technology, many microbiome studies have published, driving the emergence and advance of related analysis toolWen, Tao (Oxford University Press)
Every base everywhere all at once: pangenomics comes of age
nature.com/articles/d41586-023…
Every base everywhere all at once: pangenomics comes of age
Multi-genome assemblies called pangenomes can capture genetic diversity in a species, but researchers are still working out how best to build and explore them.Eisenstein, Michael
JASPER: A fast genome polishing tool that improves accuracy of genome assemblies
journals.plos.org/ploscompbiol…
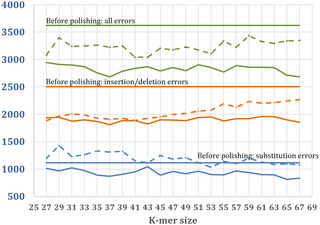
JASPER: A fast genome polishing tool that improves accuracy of genome assemblies
Advances in long-read sequencing technologies have dramatically improved the contiguity and completeness of genome assemblies.journals.plos.org
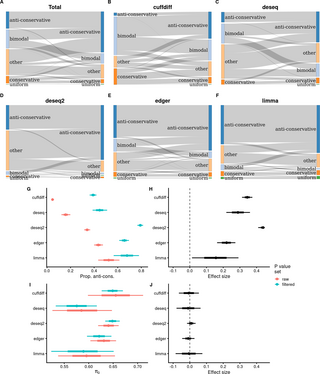
A field-wide assessment of differential expression profiling by high-throughput sequencing reveals widespread bias
A large-scale examination of differential RNA expression study datasets submitted to the NCBI GEO repository from 2008-2020 reveals widespread bias, as assessed by the distribution of p values and estimated proportions of true null hypotheses.doi.org
An overview of bioinformatics courses delivered at the academic level in Italy: Reflections and recommendations from BITS
In Italian universities, bioinformatics courses are increasingly being incorporated into different study paths.journals.plos.org
Pathogen detection in RNA-seq data with Pathonoia
bmcbioinformatics.biomedcentra…
#bioinformatics #pathogens #viruses

Pathogen detection in RNA-seq data with Pathonoia - BMC Bioinformatics
Background Bacterial and viral infections may cause or exacerbate various human diseases and to detect microbes in tissue, one method of choice is RNA sequencing.BioMed Central
biorxiv.org/content/10.1101/20…
#phylogeny #phylogenomics #bioinformatics #BigData #visualization #vizbi #myxozoan @dee_unil
1/thread
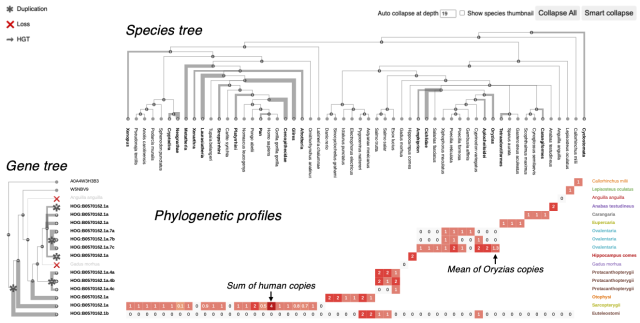
Matreex: compact and interactive visualisation of large gene families provides evidence for loss of intraflagellar transport in a myxozoan
Studying gene family evolution strongly benefits from insightful visualisations. However, the ever-growing number of sequenced genomes is leading to increasingly larger gene families, which challenges existing gene tree visualisations.bioRxiv

High-resolution metagenomic reconstruction of the freshwater spring bloom - Microbiome
Background The phytoplankton spring bloom in freshwater habitats is a complex, recurring, and dynamic ecological spectacle that unfolds at multiple biological scales.BioMed Central
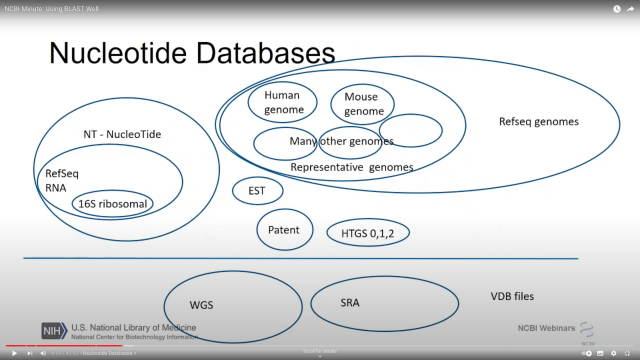
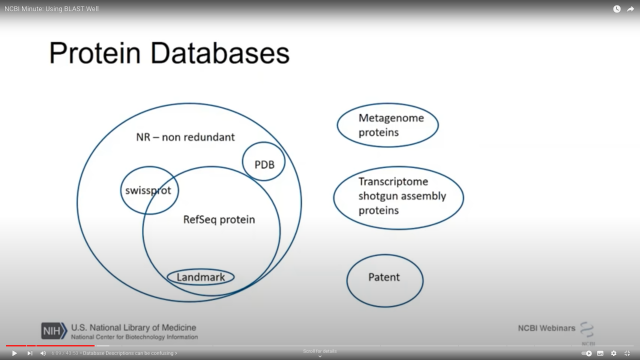
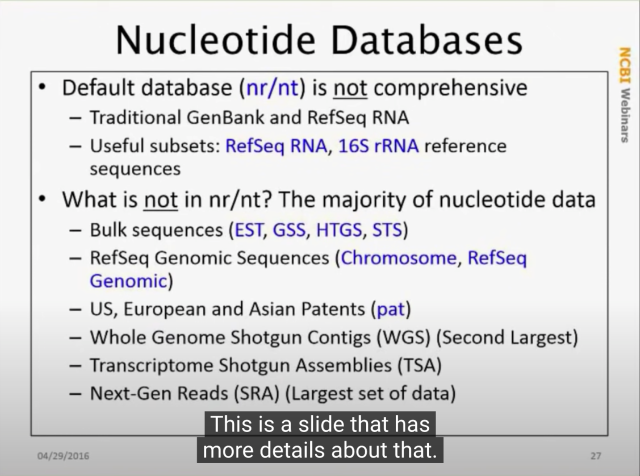
If you've ever wondered EXACTLY what is in
@NCBI
nt and nr (I certainly have!)
From youtu.be/2FW1dk5YQ3I?t=484 and youtu.be/KLBE0AuH-Sk?t=692 (cued to the right spots in the video)
NCBI Minute: Using BLAST Well
Presented October 3, 2018. In this webinar, the NCBI BLAST team lead will show you how to be more effective with NCBI’s standalone BLAST applications. You wi...YouTube
Are you passionate about #bioinformatics / #Virology / #AMR / #Microbiology / #InfectiousDisease? Another amazing faculty opportunity is opening up at the University of Toronto!
RT 😀
When I started my lab a few years ago I started building up an in-house library for students. Two of the first books I bought are The Logic of Chance (Eugene Koonin) and The Origins of Genome Architecture (Michael Lynch). Both are remarkable books that discuss genomics in the context of microbial diversity and the tree of life.
What other genomics/evolution classics do people always keep around?